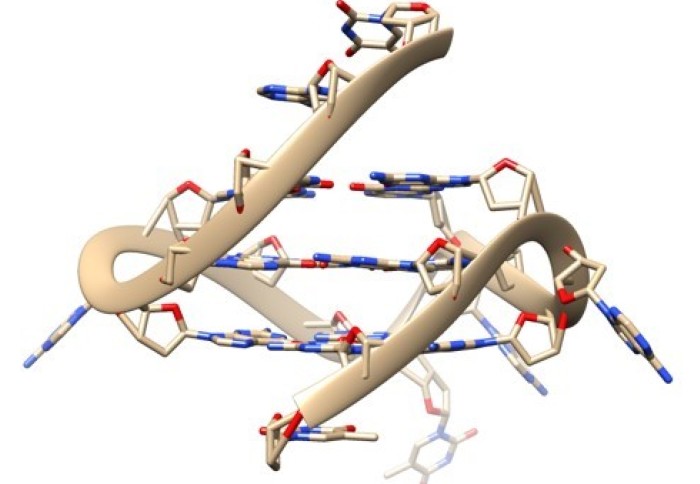
We all know the structure of DNA is a double helix ( looks like a twisted staircase). Though DNA can form some special shapes in labs, recent studies have found four-stranded DNA, known as G-quadruplex forming naturally in human cells.
Now, scientists at the Imperial College London have created new probes that can see how G-quadruplexes are interacting with other molecules inside living cells.
These four-stranded DNA also known as G-quadruplex is usually found in higher concentrations in cancer cells; hence scientists think that G-quadruplexes play a role in the disease. Further study revealed that G-quadruplexes are ‘unwound’ by specific proteins and help identify molecules that bind to G-quadruplexes, leading to potential new drug targets that can disrupt their activity.

Ben Lewis, from the Department of Chemistry at Imperial College, said that a different DNA shape will have an enormous impact on all processes which involve the DNA like reading, copying, or expressing genetic information.
We now have evidence that G-quadruplexes play an important role in a wide variety of processes vital for life, and a range of diseases, but the missing link has been imaging this structure directly in living cells.
These G-quadruplexes are uncommon inside cells . Thus standard methods for recognizing such molecules experience issues. Lewis compared the problem to finding a needle in a haystack . Further complication was that this needle was also made of a haystack.
For solving the problem analysts from the Vilar and Kuimova groups in the Department of Chemistry at Imperial collaborated teamed up with the Vannier group from the Medical Research Council’s London Institute of Medical Sciences.
They used a chemical probe called DAOTA-M2, which fluoresces (lights up) in the presence of G-quadruplexes, but instead of monitoring the brightness of fluorescence, they observed how long this fluorescence lasts. The signal from this doesn’t depend on the probe’s concentration or G-quadruplexes, so it can be used to visualize these rare molecules unequivocally.
Dr. Marina Kuimova, from the Department of Chemistry at Imperial College added that this approach helped them remove difficulties that prevented the development of reliable probes for this DNA structure.
These probes helped them study G-quadruplexes’ interaction with two helicase proteins (molecules that ‘unwind’ DNA structures) which revealed that if these helicase proteins were removed, more G-quadruplexes were formed, showing that the helicase play a role in unwinding and breaking down G-quadruplexes.
They also found out the ability of other molecules to interact with G-quadruplexes in living cells. If a molecule introduced to a cell binds to this DNA structure, it will displace the DAOTA-M2 probe and reduce its lifetime, i.e., how long the fluorescence lasts thus paving a way to study more interactions inside the nucleus of living cells and for more molecules.
Professor Ramon Vilar from the Department of Chemistry at Imperial said that the potential of G-quadruplex binding molecules as potential drugs for diseases such as cancers has immensely interested many researchers. This method will now help us in understanding the potential .
The team also claimed that it was a fantastic opportunity to work at the intersection of chemistry, biology, and physics. It would not have been possible without the expertise and close working relationship of all three research groups.
Journal Reference:
Peter A. Summers, Benjamin W. Lewis, Jorge Gonzalez-Garcia, Rosa M. Porreca, Aaron H. M. Lim, Paolo Cadinu, Nerea Martin-Pintado, David J. Mann, Joshua B. Edel, Jean Baptiste Vannier, Marina K. Kuimova & Ramon Vilar Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nature Communication 12, 162 (2021). DOI: 10.1038/s41467-020-20414-7
Press Release: Imperial College